Current methods to diagnose autism spectrum disorder rely on observing the behavior of the patient. Now, researchers are investigating metabolic pathways that may hold the clues to more definitive diagnosis.
Autism spectrum disorder (ASD) is a neuro-developmental condition characterized by difficulties in social communication and interaction, in addition to the presentation of restricted, repetitive behaviors and interests (1). The number of children diagnosed with ASD in the U.S. has increased significantly over the past 20 years. In 1996, ASD was estimated to affect one in 294 children (2); the most recent estimate is that one in 68 children is affected (3).
Treating ASD in the U.S. is estimated to cost $268 billion annually (4), and the lifetime economic burden for caring for an individual with ASD is approximately $1.4 million (5). Although a definitive cause of ASD remains elusive, many consider ASD to be caused mainly by an interaction of environmental factors in genetically predisposed individuals (6).
Due to the overall lack of understanding of the disorder’s biological origins, the current standard for diagnostic evaluation of ASD is a series of behavioral observations. However, the inherent subjectivity of a diagnosis based on observation has given rise to large variation in the age at which ASD is identified. As a result, the median age of diagnosis is approximately four years of age (3), despite stable diagnoses being possible by two years of age (7).
Identification of a set of biochemical markers for ASD to supplement the behavioral diagnosis would allow for more reliable diagnoses to be made at an earlier age. Earlier identification would allow for earlier behavioral intervention, which has been shown to improve outcomes in children, not just in the short term, but also later in life (8). Additionally, a set of biomarkers that can be used for diagnoses can also point the way to potential future treatments. With these points in mind, this article highlights some recent research from our group based at Rensselaer Polytechnic Institute that involves modeling and multivariate analysis of metabolic pathways that have the potential to serve as biomarkers for ASD diagnosis and have implications for intervention and treatment.
Metabolic pathways of interest for ASD diagnosis
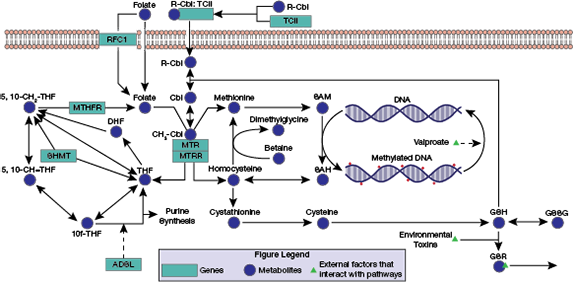
▲Figure 1. The intertwined folate-dependent one-carbon metabolism (FOCM) and transsulfuration (TS) pathways contribute to DNA methylation and the management of intracellular oxidative stress.
Two biochemical pathways of interest in the study of ASD are the folate-dependent one-carbon metabolism (FOCM) and transsulfuration (TS) pathways (Figure 1). These intertwined pathways have several roles in the human body, but two of direct interest are the epigenetic control of gene expression through DNA methylation and the management of intracellular redox status by the antioxidant glutathione (GSH).
Clinical studies of FOCM/TS pathway metabolites in individuals with ASD have consistently found a lower ratio of S-adenosyl-methionine (SAM) to S-adenosylhomocysteine (SAH), which is evidence of a lower DNA methylation capacity, as well as a lower ratio of reduced GSH to its oxidized form glutathione disulfide (GSSG), indicative of higher intracellular oxidative stress.
The Integrated Metabolic And Genomic Endeavor (IMAGE) study at Arkansas Children’s Hospital Research Institute (9) measured these and other markers of DNA methylation and oxidative stress in the plasma of children with ASD and typically developing (TD) control patients. In an effort to bridge the gap between ASD pathophysiology and clinical measurements, our research group used these data to investigate three important topics related to the disorder:
- classifying individuals with ASD, to support diagnosis
- predicting adaptive behavior in individuals with ASD, to determine severity of certain aspects of the condition
- modeling the FOCM and TS pathways in individuals with ASD, to determine the key reaction steps that are altered in children with ASD.
Classification of ASD using biochemical measurements
Biomedical researchers commonly use univariate statistical methods to compare measurements taken from two groups. However, when analyzing data from metabolic pathways such as FOCM and TS, where measurements are connected to each other via biochemical reactions, univariate methods may miss important relationships that distinguish the two groups.
Multivariate statistical techniques are better equipped to analyze these types of data, since they take into account the interactions among multiple measurements (10). In the context of a classification procedure for separating two groups of data (ASD and TD in our case), Fisher discriminant analysis (FDA) (11) is one such multivariate technique that can be used. FDA is a supervised method of dimensionality reduction that uses a linear combination of the original measurements to arrive at a single discriminant score. It does this in a way that maximizes the difference in the mean discriminant score between the ASD and TD cohorts while minimizing the within-group variance of the discriminant scores.
When identifying an FDA model for classification, it is critical to determine a subset of measurements that provides optimal separation between the two cohorts. Selecting too few measurements may not provide sufficiently useful information to separate the groups, while selecting too many can introduce unwanted noise or conflicting information into the model. A potential consequence of using too many measurements for classification is overfitting of the available data; that is, the model will describe the data used for model development exceptionally well, but will perform poorly when introduced to new data that were not used to develop the model. Cross-validation can help to alleviate concerns of overfitting by reserving subsets of data for model prediction to provide a statistically independent assessment of model performance.
A recent study by our research group (12) classified individuals into ASD and TD cohorts by applying FDA with cross-validation to data from the IMAGE study. The data included FOCM/TS plasma measurements for 83 individuals on the autism spectrum and 76 age-matched TD controls between the ages of three and ten...
Would you like to access the complete CEP Article?
No problem. You just have to complete the following steps.
You have completed 0 of 2 steps.
-
Log in
You must be logged in to view this content. Log in now.
-
AIChE Membership
You must be an AIChE member to view this article. Join now.
Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
