The complexity of the epigenome suggests a wealth of untapped biological information and regulatory potential that could be game changing for cellular engineering and biomedical interventions.
Where is information stored in humans? As engineers, we have accelerated our understanding of biology and our ability to harness it for biomedical and biotechnological applications by asking questions that provide a quantitative perspective of biological systems. We ask how much material and energy passes through metabolic pathways. We ask how much force is exerted on and by cells and tissues. We ask how strongly and how quickly genes are turned on and off in response to environmental and native signals. Correspondingly, we have developed genetic and protein tools, as well as biomaterials and fluidic devices, that can control these processes.
Although these questions are diverse, they share a common thread. We are ultimately asking how much information is stored in a biological system, where it resides, and how we can control and harness it.
One of the largest financial and human investments in biological research sought to characterize the information encoding us, through the sequencing of the human genome. A draft of the roughly 6 billion nucleic acids that make up the human genome was completed in 2003. It has been said that the human genome contains the equivalent of 1.5 gigabytes (GB) of information, as if each position within the DNA — which can be one of four nucleic acids — were represented by 2 bits of information (i.e., 00, 01, 10, 11). Yet, our intuition suggests that 1.5 GB, the size of many operating systems, computer games, and high-resolution movies, could not possibly encode the complexity of a human being. Our intuition is right.
The human body is composed of trillions of cells, and therefore, trillions of genomes. Thus, the human body could be estimated to hold over a zettabyte (1021 bytes) of information. However, each of our cells contains only copies or very similar variants of the same genomic sequence, e.g., trillions of copies of the same 1.5 GB sequence. How, then, can we explain the complexity of the human species?
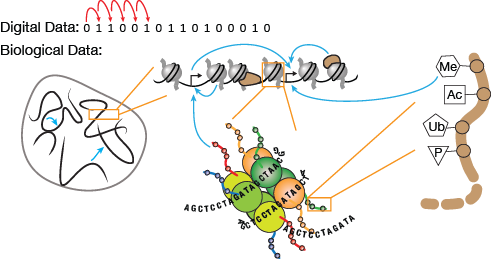
▲Figure 1. Information in digital data is read linearly (red arrows) in a bit-by-bit fashion. Information in biological systems is highly nonlinear and exhibits a complexity of layered interactions, including intra- and inter-chromosomal interactions, gene-gene regulations, and proteins and histone modifications that control genes. The blue arrows represent both physical and regulatory interactions between different components of chromatin. Blue arrows pointing to black promoter arrows signify that the particular chromatin component or feature may regulate promoter activity and gene expression. The brown backbone is the n-terminal end of one of the histone proteins.
As it turns out, information in biological systems extends well beyond the linear sequence of four nucleic acids. Biological systems do not read and write information in the same linear, one-dimensional, bit-by-bit manner that traditional transistor-based computers do. Instead, many layers of information interact with each other nonlinearly (Figure 1). This is especially true in mammalian systems, where genomes have several important properties that greatly augment their information capacity:
- mammalian genomes (as well as those of yeast and plants) are ubiquitously bound with protein complexes called nucleosomes to form beads-on-a-string-like structures
- these protein complexes can be chemically modified and have more than 60 known moieties
- hundreds of additional proteins control the spatial positioning and chemical modifications of nucleosomes
- the genome forms specific 3D structures in space.
Collectively, these features are known as the epigenome. We are still developing a deep and quantitative understanding of the information contained in the epigenome. Research shows that there are many opportunities to harness this information for biotechnological and biomedical purposes.
Why should engineers care about the epigenome? The quick answer is that the epigenome impacts almost all normal and disease processes. Thus, being able to interface with it is important for biomedical applications. A more subtle but equally important reason is that the epigenome is physically layered on the genome and thus influences when and where genes are expressed. The next sections discuss the properties of the epigenome that are important for normal cellular function and could be exploited for cellular engineering and biomedical applications.
Rheostat-like control
Gene expression levels fluctuate when specifying cell type identity or in response to environmental perturbations. Control over gene expression levels is also desired in cellular engineering applications, ranging from tuning metabolic pathway fluxes to controlling the potency of T-cell immunotherapies.
To synthetically control gene expression levels, a common strategy is to control the concentrations of proteins that bind the promoters of genes, and thus regulate their activation or repression. The epigenome provides additional...
Would you like to access the complete CEP Article?
No problem. You just have to complete the following steps.
You have completed 0 of 2 steps.
-
Log in
You must be logged in to view this content. Log in now.
-
AIChE Membership
You must be an AIChE member to view this article. Join now.
Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
