Harnessing and applying epigenetics in bioprocessing systems may improve cell line development, productivity, and product quality.
Epigenetic modifications, such as DNA methylation, chromatin modification through histone post-translational modification, and RNA interference, can have a great impact on industrial mammalian cell cultures.
For example, epigenetic factors influence several aspects of Chinese hamster ovary (CHO) strain development and scaleup. CHO cell cultures are used to produce over 70% of biopharmaceuticals, including monoclonal antibodies and other therapeutics. In CHO cell lines, integration site(s) of a monoclonal antibody gene and the method used to select for monoclonal antibody gene integration both affect long-term productivity loss during serial passages. Epigenetics is also a factor in controlling the activation of secondary metabolite products in fungal and yeast cells.
This article highlights the major modes of epigenetic-based regulation as they relate to mammalian cell culture instability and the influence of epigenetics on bioproduction in fungal and yeast systems. It also covers recent advances in engineering that enable control of the epigenetic state at a particular genomic location. It envisions how these nascent technologies may be used to improve host strain development, productivity, and product quality — focusing on the specific scientific questions that are still to be answered.
Mammalian cell culture instability
Industrial CHO cell lines are made by transfecting genes that code for therapeutic proteins (e.g., light and heavy chains for monoclonal antibodies) under the control of strong promoters (e.g., cytomegalovirus [CMV], elongation factor), which are then screened for high-producing phenotypes. CHO cells exhibit significant heterogeneity, which arises from genetic plasticity (i.e., the ability of one genotype to produce more than one phenotype) that is typical of immortalized cell cultures and cancer cells. This heterogeneity is leveraged to select for highly productive clones, but eliminates some favorable production phenotypes during scaleup and production. Note that cloning does not reduce chromosomal heterogeneity or mutation rates.
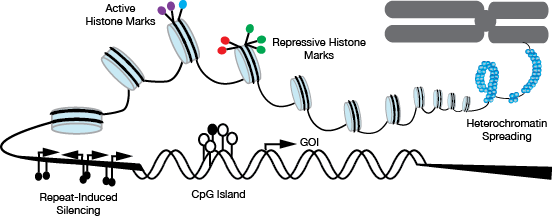
▲Figure 1. Various epigenetic processes are known to influence gene expression, degrading production productivity. These processes include heterochromatin spreading, repeat-induced gene silencing, methylation of CpG islands (white circles are methyl-free cytosine, black circles are methylated cytosine), and histone modifications. Histone modification might include loss of active histone marks — such as H3K4 methyl groups (purple circles) and H3K27 acetyl groups (blue circle). Histone modification may also entail the addition of repressive histone modifications, such as H3K27 methyl groups (green circles) and H3K79 methyl groups (red circles).
Epigenetic marks, such as histone modifications and DNA methylation, influence cells in culture in the short term and long term (Figure 1). Differences in culture conditions, such as high product and metabolic waste concentration, reactor operation mode (e.g., adherent vs. suspension, batch vs. fed batch), and media composition (e.g., serum-containing vs. serum-free, supplemented), cause transcriptional changes that are associated with genome-wide epigenetic changes.
Most studies of CHO cell instability have focused on reduction in the rate of product formation per cell (qP) over time, which manifests as a loss of mRNA due to one or a combination of several epigenetic mechanisms, including transgene genomic location, gene copy number loss, repeat-induced transcriptional silencing, and promoter methylation. The impact of these mechanisms on CHO, yeast, and fungal cell line development are covered in this article.
Introduction of a transgene into the CHO cell genome both perturbs the local chromatin structure and is influenced by the local chromatin structure. Epigenetic factors associated with the transgene can be leveraged to create and maintain a favorable chromatin structure. However, the genetic instability of the CHO cell line hinders the reliable use of transgenes.
An alternative and complementary approach is to discover regions of the genome that are favorable for transgene expression, i.e., hot spots. The genomic information necessary to identify hot spots only became available in 2011. Hot spots can be identified from stably selected clones and targeted using zinc-finger (ZF) nucleases, transcription activator-like effector nucleases (TALENs), or clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9.
Promoter methylation, which causes gene silencing and loss of gene copy number, has been examined as a cause of reduction in cell line productivity. Studies used the CMV promoter, which is most common in CHO cell transgene expression. The methylated promoter reduced productivity without a change in copy number (1, 2). A subsequent study across ten cell lines that expressed different proteins found that while promoter methylation was a factor in productivity loss, gene copy number loss was more common (3). An important difference between these studies is the selection system used (i.e., dihydrofolate reductase [DHFR] vs. glutamine synthase selection), which indicates that the particular cell line, protein, and selection system may influence the mode of epigenetic silencing. Furthermore, epigenetic silencing of the CMV promoter can occur prior to methylation through histone modifications (4), suggesting methylation may be a consequence rather than a cause of silencing.
Fungal and yeast cell cultures
CHO cells dominate the production of monoclonal antibodies and therapeutics; however, yeast and fungi can also be used to produce certain therapeutics, as well as industrial enzymes and secondary metabolites. Eukaryotic bioprocessing systems include yeast, such as Saccharomyces cerevisiae and Yarrowia lipolytica, and filamentous fungi, such as...
Would you like to access the complete CEP Article?
No problem. You just have to complete the following steps.
You have completed 0 of 2 steps.
-
Log in
You must be logged in to view this content. Log in now.
-
AIChE Membership
You must be an AIChE member to view this article. Join now.
Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
