Traditional methods of DNA synthesis are slow and costly, and hinder the design-build-test cycle for creating optimal gene sequences and protein variants. This article presents a novel approach that will allow researchers to explore synthetic biology’s full potential.
Synthetic biology is one of the most important technological advances of the 21st century. Redesigning organisms offers the potential for new breakthroughs, such as novel medicines and therapies, sustainable energy from biofuels, plastics and fibers made from sugar rather than unsustainable oil, and food crops that can fertilize themselves. The possible benefits are almost endless.
DNA reading and writing: Two faces of the same coin
In the past decade, the development of next-generation sequencing (NGS) — a fast, reliable, and affordable method of reading a DNA sequence — revolutionized synthetic biology. Using this massively parallel high-throughput method, genomes of hundreds of organisms — from bacteria and viruses to plants and animals — were sequenced, providing an unprecedented amount of genetic information with single-base resolution.
Through large-scale data curation and sharing initiatives such as GenBank and ClinGen, end-users can access genetic data, which fuels research and enables a better understanding of sequence-to-function relationships. This facilitates hypothesis-driven redesign of specific genomic loci or even whole genomes, which scientists and engineers can then test to assess the impact on, and ultimately optimize, the processes that generate useful, high-value products.
The design-build-test cycle: Accelerating evolutionary processes
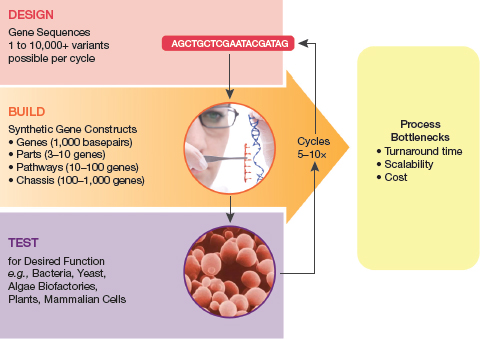
▲Figure 1. The design, build, and test cycle that underlies each DNA application development process requires DNA for rapid generation and optimization of custom pathways and organisms.
The cornerstone of synthetic biology is the design, build, and test cycle (Figure 1), an iterative process that requires DNA for rapid and affordable generation and optimization of custom pathways and organisms.
In the design phase, the adenine (A), cytosine (C), thymine (T), and guanine (G) nucleotides that constitute DNA are formulated into various gene sequences that comprise the locus or pathway of interest. Each hypothesis that will be tested requires a variant gene sequence. These variant gene sequences represent subsets of sequence space, a concept that originated in evolutionary biology and pertains to the totality of sequences that make up genes, genomes, transcriptome, and proteome.
Bioengineers compile many different variants for each design-build-test cycle to enable adequate sampling of sequence space and maximize the probability of finding an optimized design. These sequences, made accessible by DNA reading, are then manufactured through conventional synthesis methods. Once the various designs are assembled, they are tested for desired function within a chosen model system, which creates nanoscale biofactories out of cellular systems.
This cycle is repeated many times, testing permutations of the sequences and varying nucleotides at multiple positions, until an optimal candidate is identified. Although straightforward in concept, process bottlenecks related to speed, throughput, and quality slow the pace, extending development time. Despite recent advances in technologies that support the overall design-build-test process, the ability to build each hypothesis for testing remains the rate-limiting step. Bioengineers are unable to sufficiently explore sequence space due to the high cost of highly accurate DNA and the limited throughput of current synthesis technologies.
Beginning with the build phase, two processes underlie success: oligo synthesis and gene synthesis. In the past, scientists synthesized different gene variants through molecular cloning. In this process, a gene or sequence of interest is extracted from a given organism. The gene is inserted into a vector system that replicates and produces large amounts of the gene for further study. While this approach is robust, it is not scalable.
A successful design-build-test process relies heavily on the ability to iterate on the design and its underlying sequences. Supporting this cyclical process requires large amounts of DNA that consist of hundreds or thousands of sequence variants. Simply put, cycle time depends on how quickly and how broadly this DNA can be accessed for the test phase.
In William Stemmer’s classic 1995 paper dealing with the synthesis of long DNA sequences, it took approximately 50 to 60 40-mer oligonucleotides (oligos) to construct a 1-kilobase (kb) gene using assembly polymerase chain reaction (PCR) (1). Synthesis of oligonucleotides was carried out using the classic phosphoramidite chemistry developed by Marvin Caruthers (2, 3). This robust process has stood the test of time.
Most DNA synthesis platforms that exist today leverage phosphoramidite chemistry. Coupling phosphoramidite chemistry with gene assembly methods like PCR is what enables synthetic biology. New innovations centered on synthetic biology techniques improve quality, throughput, scalability, and turnaround time of bioprocesses. Technological advances in DNA writing, like those in DNA reading, drive the development of new applications in synthetic biology by increasing access to sequence space for further testing. However, unlike sequencing, DNA writing has not advanced enough to keep up with the rate at which DNA can be read, hindering the exploration of synthetic biology’s full potential.
The evolution of gene synthesis
Early chemical gene synthesis efforts focused on producing a large number of oligos with overlapping sequence homology (1). These were then pooled and subjected to multiple rounds of PCR, which concatenates the over-lapping oligos into a full-length double-stranded gene. This method of gene synthesis requires equimolar amounts of each oligonucleotide, normalization of their concentrations, and pooling of the oligonucleotides prior to gene assembly. While effective, these requirements make this method both time-...
Would you like to access the complete CEP Article?
No problem. You just have to complete the following steps.
You have completed 0 of 2 steps.
-
Log in
You must be logged in to view this content. Log in now.
-
AIChE Membership
You must be an AIChE member to view this article. Join now.
Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
