Large-scale sampling of patient microbiomes is giving one organization the tools to correlate microbiome composition with immune health, driving the deployment of safe, live biotherapeutic treatments for cancer and COVID-19.
The role of gut microbes in human health has been recognized for decades, but recent advances in technologies like next-generation sequencing, transcriptomic and proteomic analyses, and metabolic modeling have expanded our understanding of microbiome population dynamics, metabolic activity, and functional interactions with the human body.
As scientists examine microbiome composition across a broad population of individuals (1), certain microbes have been found to be associated with the presence or absence of certain disease states, including diabetes, neurological disorders, chronic kidney disease, and various cancers (2–4). Although the mechanisms of these effects are not well understood, gut microbes are known to have such diverse functions as breakdown of complex carbohydrates, conversion of dietary nutrients to bioavailable forms, maintenance of the integrity of the intestinal barrier, and production of immunostimulatory molecules (5).
In the past five years, numerous initiatives have been launched in both academia and industry to investigate the microbiome and its role. One application where significant impact has been achieved is in the manipulation of gut flora to prevent recurring intestinal Clostridium difficile infection, first through fecal microbiota transplant (FMT) and then using specific microbes as live biotherapeutic products (LBPs), the term for FDA-approved prescription probiotics. Use of LBPs to address C. difficile infection and several other gastrointestinal disorders is currently underway in various clinical trials, and numerous studies for the application of LBPs to solve more systemic diseases are at various stages.
Although low-cost, high-throughput sequencing has accelerated microbiome research and development, these studies tend to lack quantitative rigor. There is an urgent need for the use of systems biology tools like multi-omics and metabolic modeling to gain a clear mechanistic understanding of the processes by which the gut microbes impact human health (6). Much as systems and synthetic biology have revolutionized bioprocess development in the past two decades, such an engineering approach has the potential to revolutionize microbiome research and the development of LBPs.
This article discusses large-scale patient microbiome sampling and analysis of these samples in conjunction with real-world medical data to drive the development of therapeutics and diagnostics for a broad population and different diseases. A specific focus area is the impact of microbiome composition and function on immune health and how this is applied to two different therapeutic areas: cancer immunotherapy and fighting infectious viral diseases such as COVID-19.
Immunomodulatory role of the gut microbiome
One of the most important roles the gut microbiome plays is in mediating and maintaining homeostasis of the immune system. As the entry point for many pathogenic species, the intestinal region is guarded by one of the highest concentrations of immune cells — most notably B cells and T cells — in the human body. Commensal bacteria interact with these cells via surface proteins and metabolic products; such signals maintain the proper balance of activation and repression, so that the immune cells are armed in case of pathogen attack but do not produce an unnecessary inflammatory response.
For example, microbially produced short-chain fatty acids (SCFAs) like propionate and butyrate are necessary for modulation of cytotoxic T cells and maturation of regulatory T cells (7) and can regulate the production of inflammatory cytokines (8). However, such immunomodulatory signals, as well as helpful gut microbes, can be lost due to antibiotic use, chronic disease, or unhealthy lifestyle choices like poor diet, tobacco use, and alcohol use. This causes a state of dysbiosis, where normal healthy commensal microbes in the gut are reduced or lost and are often supplanted by opportunistic or pathogenic microbes that can elicit a general inflammatory response (Figure 1) (9). This can lead to autoimmune disease, diabetes, obesity, cardiovascular disease, and even neurodegenerative disorders.
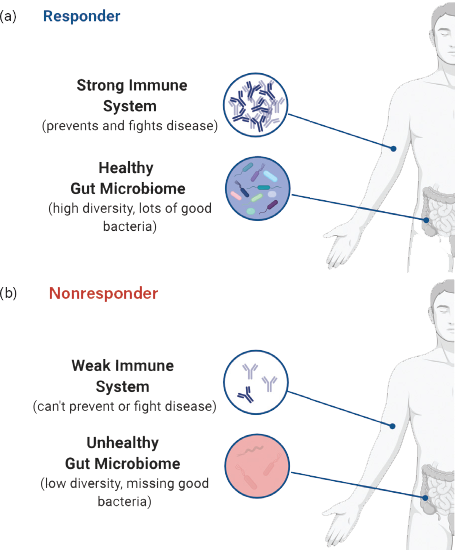
▲Figure 1. The gut microbiome modulates the immune system. (a) A healthy individual has a diverse microbiome, which produces metabolic signals to maintain a strong immune system. (b) Many commensal species are absent from a dysbiotic microbiome, which can result in a weak immune system or an inflammatory state.
AI-driven drug discovery
Persephone’s technology platform uses metagenomic sequencing and other -omics tools together with artificial intelligence (AI) and metabolic modeling to unravel the complexity of population-scale patient data (Figure 2). Rather than relying on in vitro or mouse models to identify targets, this patient-centric approach assures that the targets found are clinically relevant, and it reduces translation risk. Analysis of a diverse set of patient samples will provide confidence that the drugs developed will apply broadly to the population.

▲Figure 2. Persephone Biosciences’ technology platform for microbiome research and development begins by gathering samples from healthy and diseased patient cohorts along with medical records and metadata. Samples are analyzed with whole-genome sequencing and other -omics techniques, and artificial intelligence is used to identify microbes important for treatment efficacy.
We begin by collecting microbiome (i.e., stool) and blood samples from large populations of patients suffering from a particular disease, such as cancer, and analyzing these samples to determine microbial composition, gene expression, metabolite profile, and immune cell response. Extensive metadata, including demographics, medical history, and most importantly disease progression and therapeutic outcome, are also collected. AI is applied to these data to determine signatures of microbes, metabolites, or gene functions that differentiate a patient’s response to treatment. This allows scientists to determine the live biotherapeutics that would help restore missing microbes. Finally, several key microbial strains (generally four to six) are isolated from healthy donors and characterized. These microbes are then cultured individually by fermentation, lyophilized, and combined to formulate the LBP. The Persephone team leverages their background in the engineering of industrial bioprocesses to create a scalable manufacturing process.
One of the biggest challenges of such a large-scale data-driven approach is the actual procurement of stool samples from ill patients. While cancer patients are routinely subjected to urine and blood samplings, they are rarely asked to produce stool samples in a hospital setting. There is also a strong reluctance to provide stool samples due to the inconvenience and unpleasantness that patients associate with the process. Persephone Biosciences has developed a stool sample kit that is easy to use and minimizes unpleasant exposure to the fecal matter. Using this kit, stool is collected from cancer patients nationwide through an Institutional Review Board (IRB)-approved Poop for the Cure campaign (www.poopforthecure.us). These samples represent a broad distribution of geography, cancer types, and treatment regimens and will have long-term value as the database is built (Figure 3). Ultimately, the company will apply the stool collection and technology platform to develop live bio-therapeutic solutions for various indications, general health, and disease prevention.
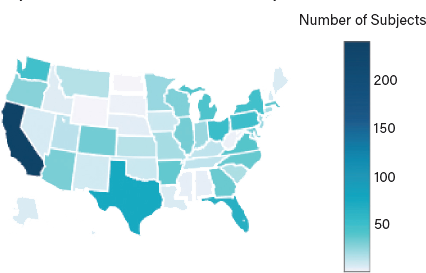
▲Figure 3. Poop for the Cure registered donors are broadly distributed in geographic location, as well as cancer types and treatment regimens.
Involvement of the gut microbiome in cancer treatment
Cancer is the second leading cause of death in the U.S., with associated healthcare costs for diagnosis and treatment exceeding $125 billion per year (10). In recent decades, immunotherapies that prompt a person’s immune system to attack cancerous cells have shown great promise for treating several types of cancers, including melanoma, lung cancer, and head and neck cancer (10). The most clinically advanced immunotherapy drugs are checkpoint inhibitors, which are monoclonal antibody drugs that target and block inhibitory regulatory pathways in T cells, thereby enabling antitumor immune responses (11).
Examples of T-cell inhibitory signal targets and their corresponding immunotherapy agents include:
- cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), targeted by, e.g., Yervoy (ipilimumab)
- the programmed cell death protein 1 (PD-1), targeted by, e.g., Keytruda (pembrolizumab) and Opdivo (nivolumab)
- the ligand PD-L1 on PD-1, targeted by, e.g., Tecentriq (atezolizumab), Bavencio (avelumab), and Imfinzi (durvalumab).
Blockade of these inhibitory signals by checkpoint inhibitor immunotherapies has been shown to be particularly effective against advanced melanoma, non-small cell lung cancer, and renal cell carcinoma, yet more than 50% of cancer patients subjected to checkpoint inhibitor therapies fail to respond to the treatment (12). These treatments are often the last line of defense, so failed response to checkpoint inhibitors is often terminal. Thus, there is a significant unmet need for improving the efficacy of immunotherapy drugs.
Overwhelming evidence has shown that nonresponsiveness to immunotherapy can be linked to a patient’s microbiota. Recently, analysis of cancer patient samples identified a strong correlation between abundances of certain microbial species in the gut and positive response to checkpoint inhibitor therapy (13–17). As a way to demonstrate that the gut microbiome impacts response to checkpoint inhibitor therapy, researchers performed fecal microbiota transplants (FMTs) from responding and nonresponding cancer patients into mice to see if the mice would have a similar response to therapy as the patient FMT donors. One study found that the mice that received a FMT from a responding patient experienced more tumor mass reduction than the mice that received a FMT from a nonresponding patient (15). These and other studies also link certain commensal microbes to increases in cytotoxic T cells and decreases in T regulatory cells and suppressive myeloid cells, indicating a more robust antitumor response; therefore, a healthy and intact gut microbiome is a necessary condition for checkpoint inhibitor function.
Development of LBPs to improve the efficacy of checkpoint inhibitors is the first application of Persephone’s technology platform. Through Poop for the Cure and clinical collaborations, stool samples are collected from immunotherapy patients suffering from a variety of different cancers to identify microbes that can increase a patient’s immunologic ability to respond to checkpoint inhibitor therapy.
As described previously, AI is applied to these data to determine signatures of microbes, metabolites, or gene functions that differentiate patients who do well on treatment (responders) from those who show disease progression (nonresponders). In general, responders have diverse microbiomes like those of healthy individuals, while nonresponders have lower diversity and are missing key microbes (Figure 4).

▲Figure 4. Patients who do well on treatment (responders, center) have diverse microbiomes like those of healthy individuals (right). Each differently shaded wedge represents the abundance of a single microbial species. The nonresponder (left) has a less-diverse microbiome.
Analysis of hundreds of samples has confirmed this general pattern. This forms the basis of a diagnostic method to predict response in advance, as well as live biotherapeutics that can be introduced to restore the missing microbes. Use of real-world patient data to generate hypotheses in this manner will ultimately reduce translational risk for entry into the clinic.
Nonclinical animal experiments were performed to validate the hypotheses generated from the data-driven approach and to test an initial LBP formulation generated from the data. Mouse cancer models (CT26, colon cancer) were administered anti-CTLA-4 checkpoint inhibitor, either alone or in conjunction with an LBP derived from Persephone’s data. Mice receiving anti-CTLA-4 alone had a modest response, reducing the tumor volume on average about 50% relative to untreated mice. In contrast, mice receiving both anti-CTLA-4 and the LBP exhibited a nearly tenfold reduction in average tumor volumes (Figure 5). Interestingly, the LBP alone reduced tumor growth nearly equal to that of the checkpoint inhibitor alone.
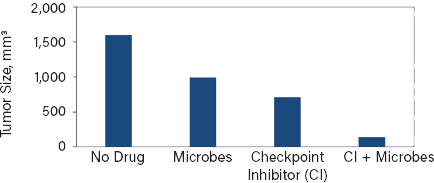
▲Figure 5. Tumors in mice receiving a checkpoint inhibitor alone shrank to about half the size of the tumors in untreated mice. In contrast, mice receiving both inhibitor and the microbe exhibited a nearly tenfold reduction in average tumor volumes. Interestingly, the microbe alone also reduced tumor growth.
The combination therapy reduced the variability in response — all mice tested were classified as responders based on final tumor volume. Variability is the primary weakness of immunotherapy; these experiments demonstrate the potential of a LBP co-therapy to improve the consistency of treatment outcome.
The gut microbiome and resistance to viral infections
The COVID-19 global pandemic has driven a race for treatment options. Current efforts have focused on using repurposed antiviral drugs, antibodies, and vaccines specific to the SARS-CoV-2 virus. While these may be short-term solutions, they alone are not enough to help patients with severe life-threatening complications.
Incidences of severe COVID-19 disease symptoms such as acute respiratory distress syndrome (ARDS) and cytokine storm (a severe immune reaction in which the body releases too many cytokines into the blood too quickly) have been more prevalent in individuals who are elderly and those with underlying health issues, such as diabetes, hypertension, autoimmune disease, cancer, cardiovascular disease, and obesity (18). All of these diseases are linked to inflammation and dysregulation of the immune system.
Studies have demonstrated the important role of the gut microbiome in maintaining immunological homeostasis, protecting against viral infections, and improving the efficacy of vaccinations (19–21). Recent reports from China suggest that COVID-19 is strongly linked to the gastrointestinal tract (22) — patients with a dysbiotic or damaged gut microbiome cannot mount an effective immune response to fight the virus, even when vaccinated or treated with antiviral medications, and thus are more likely to have longer lasting disease and severe complications.
Recent work has also suggested that certain genera of gut microbes are associated with either mild or advanced COVID-19 disease states (23). This microbiome-mediated immune stimulation is the same mechanism required by cancer patients to respond effectively to immunotherapy drugs. Therefore, we propose using the same LBP formulations described earlier as a prophylactic or adjuvant therapy against SARS-CoV-2 and other infectious viruses such as influenza.
Persephone Biosciences proposes a two-pronged approach to restoring gut microbiome function to enable the immune system to fight viral infections (Figure 6). The first is as a vaccine adjuvant (administered several days prior to the vaccine), aimed at improving the probability of a person successfully responding to a vaccine. Vaccine efficacy rates for respiratory viruses, such as influenza and SARS-CoV-2, have been historically poor, particularly for vulnerable populations such as the elderly and those with autoimmune disease, diabetes, or hypertension — the same population more likely to have a dysbiotic gut microbiome.
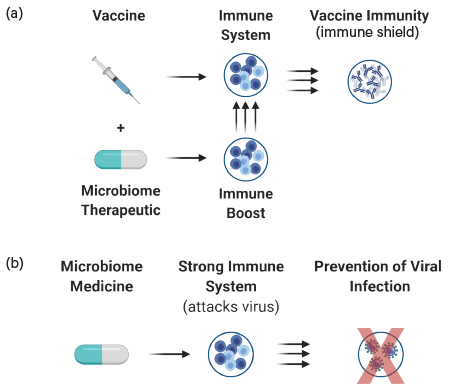
▲Figure 6. A two-pronged approach to microbiome-based therapies could be effective at fighting viral infections such as COVID-19. (a) A vaccine adjuvant creates an immune shield that increases the success rate for immunization. (b) Administering the LBP as a prophylactic will stimulate the immune cells to respond to and eliminate the virus before serious disease develops.
Adjuvants are small molecules, proteins, or other ingredients that help the body produce an immune response strong enough to protect the person from the disease he or she is being vaccinated against. These function by boosting the immunogenicity of the vaccine antigen or providing chemical signals that modulate the immune system. Various adjuvants have been used in conjunction with influenza vaccine, though efficacy rates are still low, often less than 40% (24). Our LBP adjuvant will prime the gut microbiome for optimal stimulation of the immune system to make antibodies once the vaccine is administered.
At the onset of a new global pandemic such as COVID-19, vaccines targeting the novel virus have not yet been developed. Societal response to the outbreak aims to slow the spread of the virus and to reduce the severity of the disease. Persephone’s second approach is to administer the LBP as a probiotic, conditioning the microbiome to best enable the individual’s immune system to eliminate the virus rapidly upon infection. This can occur by various mechanisms, such as increasing the ability of the immune system to combat infection, stimulate activity of specific classes of immune cells, provide essential nutrients that may be depleted or blocked by the virus, or produce compounds with antiviral activity.
The LBP would be taken either prophylactically or at the first sign of infection, much as one might take zinc lozenges, vitamin C, or Echinacea as an immune booster. This would allow the patient to clear the virus and recover more quickly from the infection, reduce the severity of disease, and lessen the likelihood of morbidity or mortality. Unlike vaccines or other therapies, which must be modified as the virus mutates (e.g., influenza vaccines are new every year), this LBP is a general immune enhancer that does not depend on the nature of the virus. Thus, use of such a treatment early can significantly curtail the spread of future pandemics.
The future of microbiome-based medicines
Persephone’s novel technology platform has the potential to revolutionize the microbiome field, as well as the entire approach to drug development. Traditionally, candidate drugs are discovered using a reductionist approach. A mechanism is first demonstrated at the molecular level in vitro; if it is successful, it is then tested in animal models and finally in humans. A significant risk is translation — drugs with binding activity or even with efficacy in mice often fail in clinical trials. In our approach, the evidence for activity comes directly from human data, prior to evaluation in vitro or in animal models, so there is less risk during the translation from preclinical to clinical studies. Furthermore, the fact that these microbes are ubiquitous in healthy human guts significantly reduces safety risk.
Persephone’s stool bank and database were developed for oncology applications, but the platform is versatile and can be leveraged to create diagnostics and therapeutics for any disease. As the database grows with data from patients with various diseases as well as healthy controls, our overall understanding of the connection between microbiome composition and the immune system also improves. As evidenced by the company’s rapid expansion of its methodology from cancer to COVID-19, having access to this data can greatly accelerate new therapeutic programs.
Literature Cited
- Turnbaugh, P. J., et al., “The Human Microbiome Project,” Nature,449 (7164), pp. 804–810 (2007).
- Flint, H. J., et al., “The Role of the Gut Microbiota in Nutrition and Health,” Nature Reviews Gastroenterology & Hepatology,9 (10), pp. 577–589 (2012).
- Pan, W., and Y. Kang, “Gut Microbiota and Chronic Kidney Disease: Implications for Novel Mechanistic Insights and Therapeutic Strategies,” International Urology and Nephrology, 50 (2), pp. 289–299 (Aug. 2017).
- Qin, J., et al., “A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes,” Nature, 490 (7418), pp. 55–60 (2012).
- Kau, A. L., et al., “Human Nutrition, the Gut Microbiome and the Immune System,” Nature, 474 (7351), pp. 327–336 (2011).
- Xavier, J. B., et al., “The Cancer Microbiome: Distinguishing Direct and Indirect Effects Requires a Systemic View,” Trends in Cancer, 6 (3), pp. 192–204 (Mar. 2020).
- Luu, M., et al., “Regulation of the Effector Function of CD8+ T cells by Gut Microbiota-Derived Metabolite Butyrate,” Scientific Reports,8 (14430) (2018).
- Schirmer, M., et al., “Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity,” Cell,167 (4), pp. 1125–1136 (2016).
- Zeng, M. Y., et al., “Mechanisms of Inflammation-Driven Bacterial Dysbiosis in the Gut,” Mucosal Immunology,10 (1), pp. 18–26 (2017).
- Mariotto, A. B., et al., “Projections of the Cost of Cancer Care in the United States: 2010-2020,” Journal of the National Cancer Institute,103 (2), pp. 117–128 (2011).
- Sharma, P., and J. P. Allison, “The Future of Immune Checkpoint Therapy,” Science,348 (6230), pp. 56–61 (2015).
- Ribas, A., and J. D. Wolchok, “Cancer Immunotherapy Using Checkpoint Blockade,” Science,359 (6382), pp. 1350–1355 (2018).
- Gopalakrishnan, V., et al., “Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients,” Science,359 (6371), pp. 97–103 (2018).
- Routy, B., et al., “Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors,” Science, 359 (6371), pp. 91–97 (2018).
- Vétizou, M., et al., “Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota,” Science,350 (6264), pp. 1079–1084 (2015).
- Iida, N., et al., “Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment,” Science,342 (6161), pp. 967–970 (2013).
- Matson, V., et al., “The Commensal Microbiome is Associated with Anti-PD-1 Efficacy in Metastatic Melanoma Patients,” Science,359 (6371), pp. 104–108 (2018).
- Guo, Y.-R., et al., “The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak — An Update on the Status,” Military Medical Research,7 (11) (2020).
- Li, N., et al., “The Commensal Microbiota and Viral Infection: A Comprehensive Review,” Frontiers in Immunology,10 (1551), doi:10.3389/fimmu.2019.01551 (July 2019).
- Ichinohe, T., et al., “Microbiota Regulates Immune Defense Against Respiratory Tract Influenza A Virus Infection,” Proceedings of the National Academy of Sciences, 18 (13), pp. 5354–5359 (2011).
- Hagan, T., et al., “Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans,” Cell,178 (6), pp. 1313–1328.e13 (2019).
- Gao, Q. Y., et al., “2019 Novel Coronavirus Infection and Gastrointestinal Tract,” Journal of Digestive Diseases,21 (3), pp. 125–126 (Mar. 2020).
- Gou, W., et al., “Gut Microbiota May Underlie the Predisposition of Healthy Individuals to COVID-19,” medRxiv, doi:10.1101/2020.04.22.20076091 (Apr. 2020).
- Tregoning, J. S., et al., “Adjuvanted Influenza Vaccines,” Human Vaccines & Immunotherapeutics, doi:10.1080/21645515.2017.1415684 (2018).

Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
