(553d) Discovery of Antibodies Targeting Multipass Transmembrane Proteins Using a Suspension Cell-Based Evolutionary Platform
AIChE Annual Meeting
2023
2023 AIChE Annual Meeting
Food, Pharmaceutical & Bioengineering Division
Engineering Protein Therapeutics
Wednesday, November 8, 2023 - 1:24pm to 1:42pm
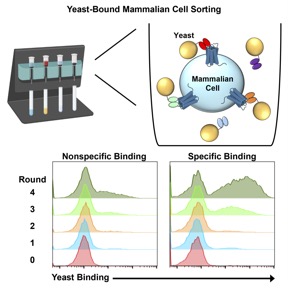
Building on the biofloating platform, we developed an optimized workflow for the selection and screening of antibodies targeting membrane proteins from naïve libraries. We also established the first reported stoichiometric binding metric to enable real-time tracking of enrichment progress and inform decisions regarding selection scheme and individual clone screening. We implemented our selection workflow against the single-pass transmembrane protein programmed death-ligand 1 (PD-L1) and discovered three unique target-specific antibody fragments. We further tested our platform against complex, multipass membrane proteins in the GPCR family, and discovered single-domain antibody (nanobody) binders against 4 different proteins: (1) C-X-C motif chemokine receptor 2 (CXCR2); (2) glucagon-like peptide 1 receptor (GLP1R); (3) glucagon receptor (GCGR); and (4) C-X-C motif chemokine receptor 4 (CXCR4). Interestingly, a discovered nanobody against CXCR2 was found to be non-competitive with the IL-8 ligand and previously reported antibodies targeting the N-terminus of the GPCR, suggesting that our evolved molecule binds a unique epitope on the receptor. Finally, we used the biofloating platform to discover antibodies against a ligand-gated ion channel that is aberrantly expressed in leukemia cells. Overall, these studies highlight the robustness and versatility of our novel approach to the discovery of antibodies targeting both single-pass and multipass transmembrane proteins for a broad range of basic and translational research applications.