(618f) Molecular Mechanisms behind Myc Regulation and Function in Cancer
AIChE Annual Meeting
2023
2023 AIChE Annual Meeting
Food, Pharmaceutical & Bioengineering Division
Systems and Quantitative Biology: Disease Mechanisms, Biomarkers, and Therapies I
Sunday, November 5, 2023 - 5:00pm to 5:18pm
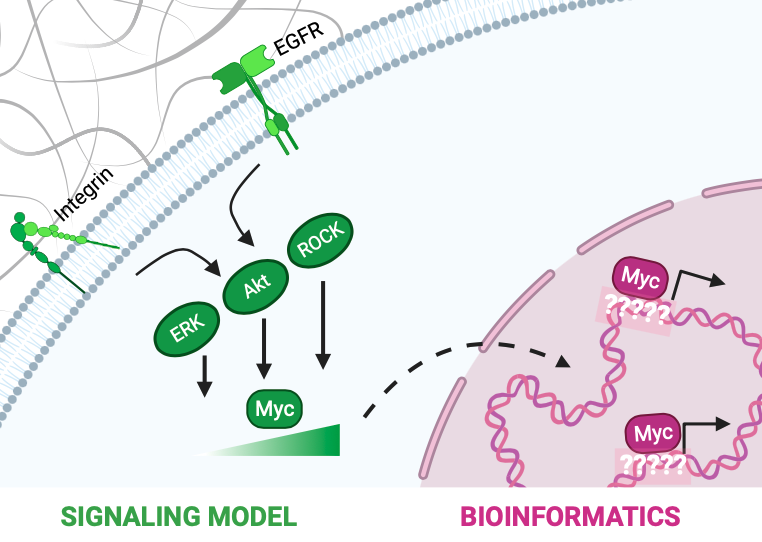
The transcription factor Myc is known to modulate a multitude of genes and cellular processes. Myc is deregulated in 70% of human cancers and is commonly known as an “undruggable†molecule. Factors such as Myc’s structure and primary nuclear localization make it difficult to directly target Myc in cancer treatments. This has led to an increased interest in identifying indirect ways to target Myc. Therefore, in this work, we sought to understand the molecular mechanisms behind 1) regulation (at the signaling level) and 2) function (at the transcription level) of Myc. In the first part of this work, we used an ODE-based kinetic modeling approach to study how extracellular mechanical and growth cues transduced through intracellular signaling pathways can drive Myc protein stabilization and accumulation in cells. The function of Myc in cancer is well-studied, however, the role of mechanotransduction in driving and sustaining cancers through Myc is yet to be uncovered. Understanding this is important as tissue stiffening is a precursor of various solid cancers. Therefore, we modeled Myc phosphorylation by MAPK, Rho/ROCK, and PI3K/Akt signaling pathways effected by EGF and integrin receptors. Our modeling results show that for normal cellular phenotypes, signaling through the EGF receptor strongly influences Myc phosphorylation. However, in cancerous phenotype, signaling through both EGF and integrin receptors play a combined role in modulating Myc. We, therefore, conclude that growth and mechanical signals play a synergistic role in driving and sustaining cancers by modulating Myc levels. Based on this, we propose that tissue stiffness must be considered as a factor while developing treatment strategies for Myc-driven solid cancers. Further, we systematically perturbed signaling interactions in the model to identify key processes governing Myc in cells. From this analysis, we identified intermediate nodes critical for Myc deregulation, which can be used as indirect targets to treat Myc-driven solid cancers. In the second part of this work, we performed bioinformatics analysis on Myc ChIP-seq datasets to study Myc-DNA interactions. Through motif discovery analysis, we identified a new DNA binding sequence of Myc which we propose is a low-affinity binding site for Myc. From subsequent bioinformatics analysis, we demonstrate that Myc functions cooperatively with other proteins to occupy this low-affinity site and drive transcription of genes. Our future work includes using CRISPR technology and live-imaging of single cells to experimentally complement the bioinformatics work and study the kinetics behind transcription by Myc. The DNA-binding patterns and protein/protein interactions identified from this work will aid in the development of cancer treatment strategies targeting Myc. Overall, in this work, we bridge signaling and transcription to gain a mechanistic understanding of Myc regulation and function in cells. We also identify new interactions and mechanisms critical to Myc activity, which can be used as indirect ways to target Myc in cancer therapies.