(463d) Soft Sensing of Intracellular States for Online Monitoring of Product Quality Using Ensemble Kalman Filters and Recurrent Neural Networks
AIChE Annual Meeting
2023
2023 AIChE Annual Meeting
Computing and Systems Technology Division
Industrial applications in Design and Operations II
Thursday, November 9, 2023 - 1:33pm to 1:54pm
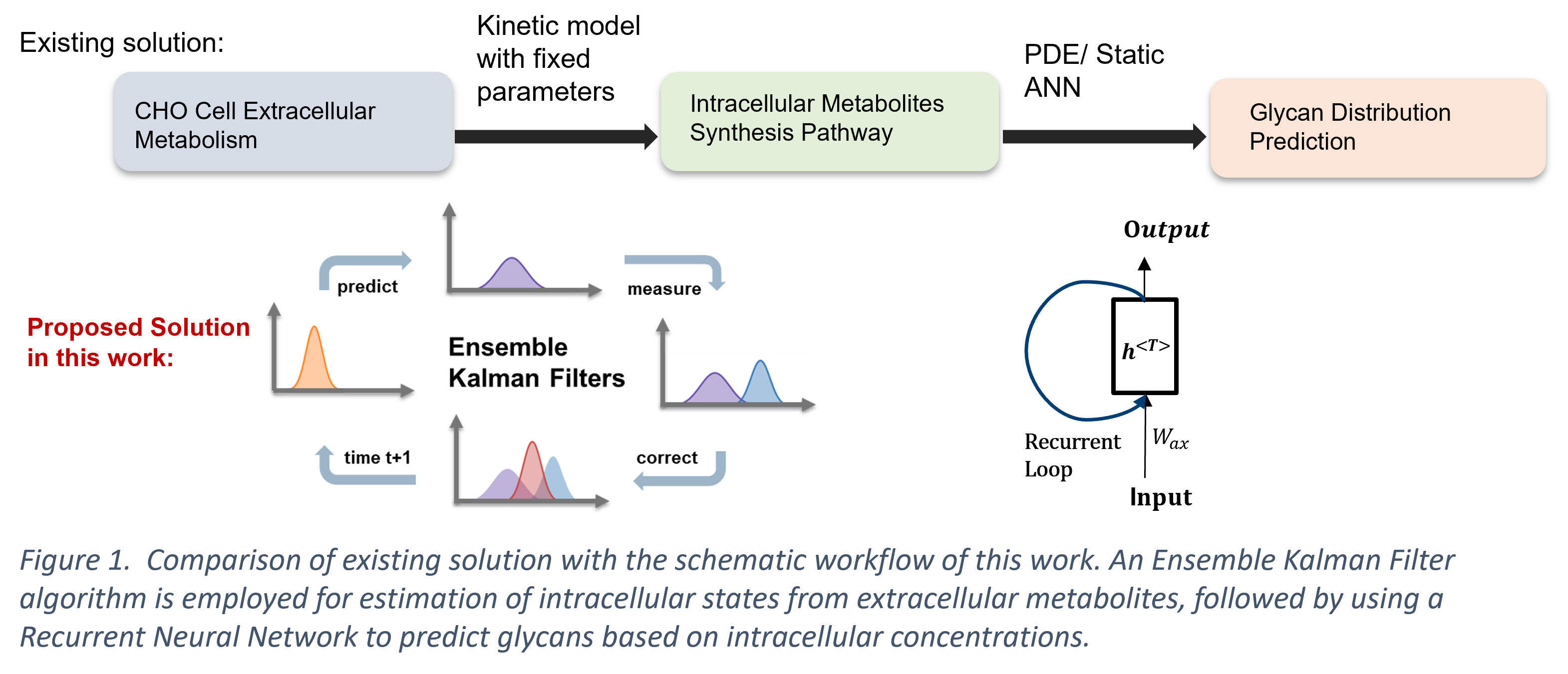
In this work, we present a modelling framework that estimates the intracellular states with an Ensemble Kalman Filter (EnKF) soft sensor, followed by online prediction of the glycan distribution through the estimated intracellular concentrations with a Recurrent Neural Network (RNN). Typically, soft sensors based on first-principles models are specifically curated for the system, limiting their applicability in online monitoring and control purposes. The EnKF combines the mechanistic predictive model with the accessible secondary extracellular measurements for estimation of the primary intracellular states, which mitigates the process variability with a probabilistic manner (Yuan et al., 2017). Herein, the EnKF addresses the issue of high specificity of the mechanistic kinetic model, while it makes use of the limited discrete sensor observations that would be otherwise unsuitable for training a standalone data-driven model. Additionally, the use of RNNs for predicting the glycan distribution is proposed in this work as an alternative to the existing PDE (Jimenez del Val et al., 2011; Kotidis et al., 2019) or ‘static’ feed-forward neural network approach (Kotidis & Kontoravdi, 2020). RNNs are able to account for the time-dependent sequential process of glycosylation, capturing the hidden dynamic relationship without acquiring the complex enzyme kinetics. The presented hybrid framework consisting of the EnKF and the RNN algorithm demonstrates greater flexibility compared to a first-principles model without sacrificing the biological understanding of the system. The proposed framework serves as a basis towards online monitoring and control of key product quality attributes such as glycosylation.
References
Jimenez del Val, I., Nagy, J. M., & Kontoravdi, C. (2011). A dynamic mathematical model for monoclonal antibody N-linked glycosylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnology Progress, 27(6), 1730–1743.
Jose De Assis, A., & Maciel Filho, R. (2000). Soft sensors development for on-line bioreactor state estimation. In Computers and Chemical Engineering (Vol. 24).
Kotidis, P., Jedrzejewski, P., Sou, S. N., Sellick, C., Polizzi, K., del Val, I. J., & Kontoravdi, C. (2019). Model-based optimization of antibody galactosylation in CHO cell culture. Biotechnology and Bioengineering, 116(7), 1612–1626.
Kotidis, P., & Kontoravdi, C. (2020). Harnessing the potential of artificial neural networks for predicting protein glycosylation. Metabolic Engineering Communications, 10.
Luo, Y., Kurian, V., & Ogunnaike, B. A. (2021). Bioprocess systems analysis, modeling, estimation, and control. In Current Opinion in Chemical Engineering (Vol. 33). Elsevier Ltd.
Yuan, X., Wang, Y., Yang, C., Gui, W., & Ye, L. (2017). Probabilistic density-based regression model for soft sensing of nonlinear industrial processes. Journal of Process Control, 57, 15–25.