(197an) Adsorption of Adeno-Associated Virus Onto Ion-Exchange Chromatographic Media: A Coarse-Grained Molecular Dynamics Study
AIChE Annual Meeting
2023
2023 AIChE Annual Meeting
Computational Molecular Science and Engineering Forum
Poster Session: Computational Molecular Science and Engineering Forum
Monday, November 6, 2023 - 3:30pm to 5:00pm
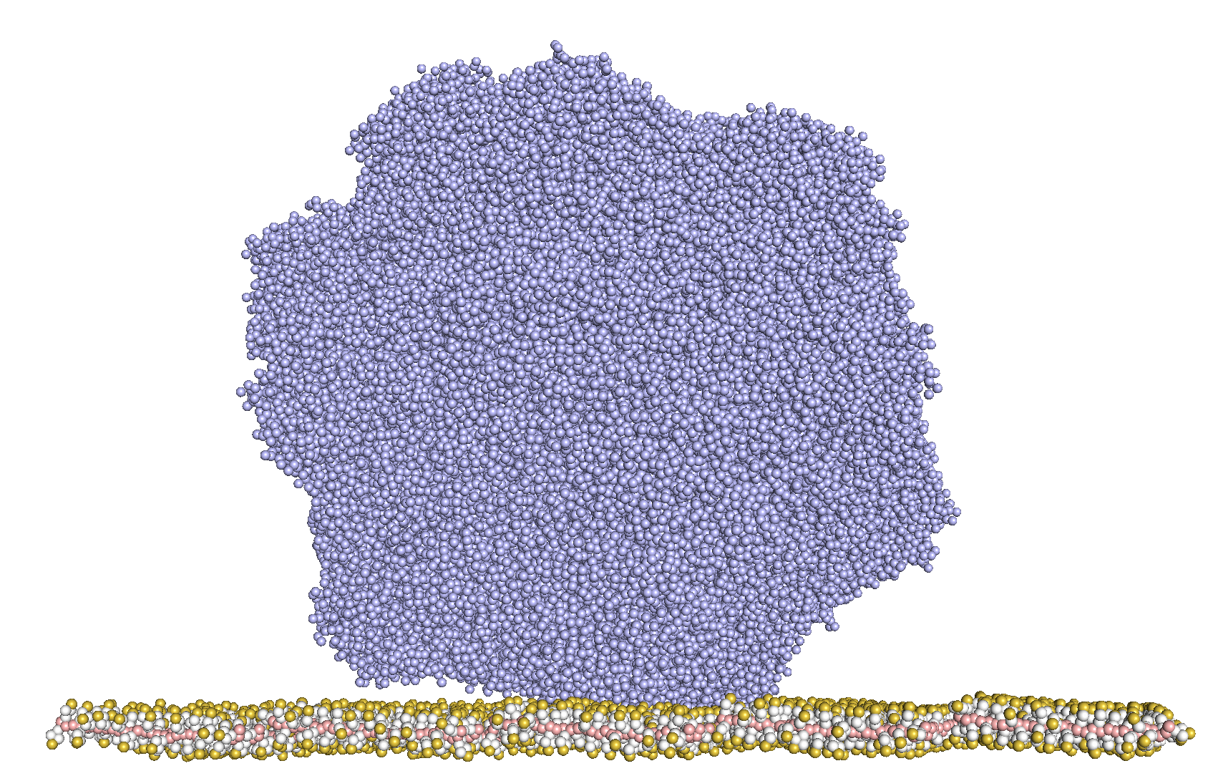
Adeno-associated virus (AAV) packaging a single-stranded (ssDNA) genome has emerged as a promising vector and is the most common vehicle for in vivo gene transfer. They are well established in clinical trials for in vivo gene therapy with a good safety profile, low immunogenicity, and long-term stability. Currently, there is limited understanding regarding the mechanism involved in using anion-exchange chromatography to separate empty and full AAV capsids under different pH conditions during purification, as well as the capsids' propensity for adsorption onto the resin material and the role of loaded ssDNA in the adsorption process. Therefore, a coarse-grained molecular dynamics (CG-MD) simulations was conducted to probe the underlying mechanism of the adsorption and interactions between the capsid and strong anion exchange resin, such as polystyrene quaternary ammonium.
Methods:
The AAV Serotype 8 (AAV8) was chosen as the model viral capsid (PDB: 6v12) with loaded 2kb ssDNA built using the Avogadro software. The initial resin material was constructed and parameterized to the coarse-grained MARTINI force field and model. All the CG-MD simulations were conducted using the GROMACS software package and the trajectories were extracted and analyzed. The steepest descent algorithm was used in energy minimization followed by a 10 ns equilibration step in an isothermal−isochoric ensemble at ambient temperature. The simulation production runs were performed in an isothermal-isobaric ensemble using the NoseÌ−Hoover thermostat and the Parrinello−Rahman barostat at a pressure of 1 bar for each 10-fs time step. The simulations were carried out using periodic boundary conditions and performed computations in the High Performance Center Supercomputer Cluster at the University of Connecticut.
Results:
The adsorption behavior of capsid onto the anion-exchange chromatography resin surface at neutral and basic solution conditions was successfully determined using the CG-MD simulations. The analysis of the center-of-mass distance demonstrated that the full capsid bound to the surface more rapidly under basic conditions than other conditions. Conversely, empty capsids did not exhibit any adsorption under neutral conditions. Furthermore, the adsorption process was found to involve multiple asparagine and glutamine as the first amino acid residues to interact with the surface. Among the various conditions tested, the full capsid under higher temperature and basic conditions exhibited the highest level of instability. This instability was indicated by the higher root-mean-square deviation (RMSD) values and significant structural changes when binding to the resin, evidenced by the relative shape anisotropy. In addition, the full capsid under basic conditions had approximately 6% more anionic charge on its surface and exhibited much stronger binding interaction energy with the surface compared to the full capsid under neutral conditions.
Conclusions:
The CG-MD simulations enhanced our understanding of the adsorption mechanism between the AAV capsids and the anion-exchange chromatography. The developed computational model allowed us to evaluate the stability, electrostatic charges, and interaction energy of the empty and the ssDNA-loaded full capsids. Our simulations offered dynamic insights into the structure and energy of the capsids on a microsecond timescale, which revealed behavior that is unattainable through experimental investigations. The CG-MD simulations are demonstrated to be a powerful tool in the simulation of macromolecular complexes capable of guiding the interpretation, explanation, and direction of experiments.