2020 Virtual AIChE Annual Meeting
(52c) An in Vitro Platform Using DNA Handles to Spatiotemporally Control Multiple Bioactive Peptides
Authors
METHODS: Norbornene-modified HA (NorHA) hydrogels were synthesized by dissolving NorHA in a photoinitiator and crosslinked with nondegradeable DL-dithiothreitol (DTT) when exposed to UV light. The total molar equivalence with DTT was set to 20% leaving the remaining 80% of the reaction sites available for tethering ssDNA; the hydrogels were soaked in a ssDNA/photoinitiator solution followed by photoconjugation using a photomask for spatial control and rinsed. A complementary fluorescently labeled biomolecule strand was added, rinsed, and imaged. For temporal control, the biomolecule strands and the complementary displacement strands were designed with a toe-hold region allowing for the removal of the biomolecule strands from the hydrogel surface. Using this approach, DNA handles were used to spatiotemporally tether peptides of interest, including RGD for cell-matrix adhesion, HAVDI for cell-cell adhesion, and OGP for osteogenesis. Cell behavior on these hydrogel surfaces were assessed using a phalloidin stain to analyze cell morphology and alkaline phosphatase staining as an early marker of osteogenesis.
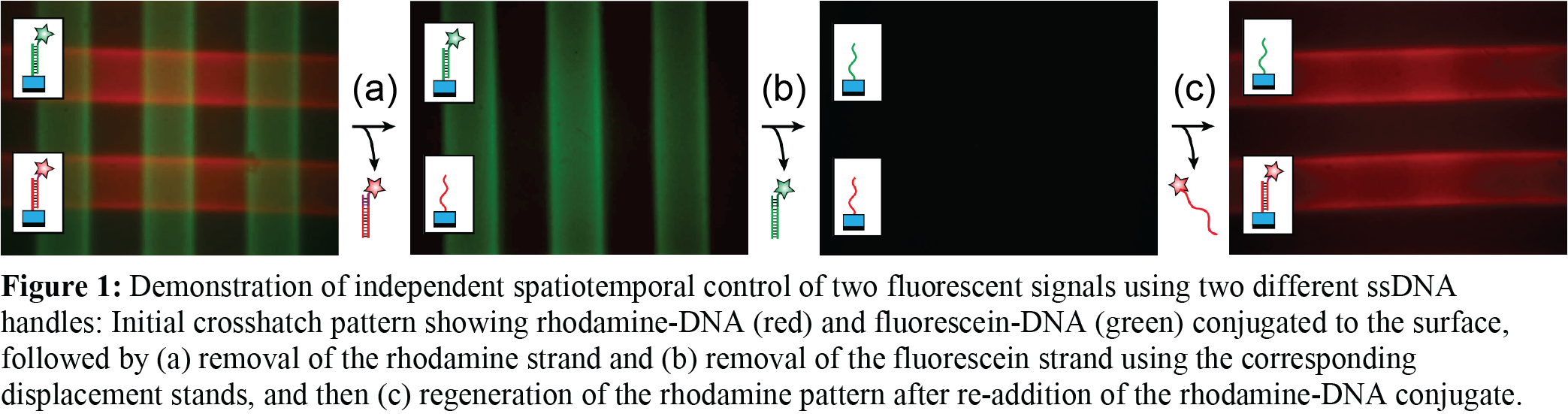
RESULTS AND DISCUSSION: To spatially and temporally control bioactive peptide delivery for improved control over cell behavior, ssDNA was bound to the surface of NorHA hydrogels through photoconjugation using a photomask with 200 μm thick lines. When complementary biomolecule strands were added, 200 μm thick green fluorescent lines were observed demonstrating that DNA was spatially bound to the surface. After adding a complementary displacement strand, no fluorescent lines were detected demonstrating complete removal of the biomolecule strand from the surface. Mismatched (non-complementary) biomolecule strands, mismatched displacement strands, and having no DNA bound to the hydrogel surface served as controls and resulted in no fluorescent activity. Hydrogels were also made with two different ssDNA handles photopatterned to the surface resulting in two different complementary biomolecule strands spatially arranged in a crosshatch pattern, Figure 1. One of the two biomolecule strands was removed and re-added to show precise temporal control. This work demonstrates that using NorHA as the platform allows for spatial control, ssDNA handles are required to be bound to the hydrogel surface for biomolecule attachment, and that the correct DNA sequence on the biomolecule and displacement strands is required for proper temporal control. Fluorescent molecules were used as a model while ongoing work is using complementary peptide-conjugated ssDNA strands to evaluate the complicated interplay between cell adhesion (both cell-cell and cell-matrix) and osteogenesis.