(153g) A Low-Cost Strategy for Genotypic Antimicrobial Resistance Detection Using Oligonucleotide Ligation Assay
AIChE Annual Meeting
2022
2022 Annual Meeting
Topical Conference: Chemical Engineers in Medicine
Chemical Engineering Principles Advancing Medicine I
Monday, November 14, 2022 - 2:36pm to 2:57pm
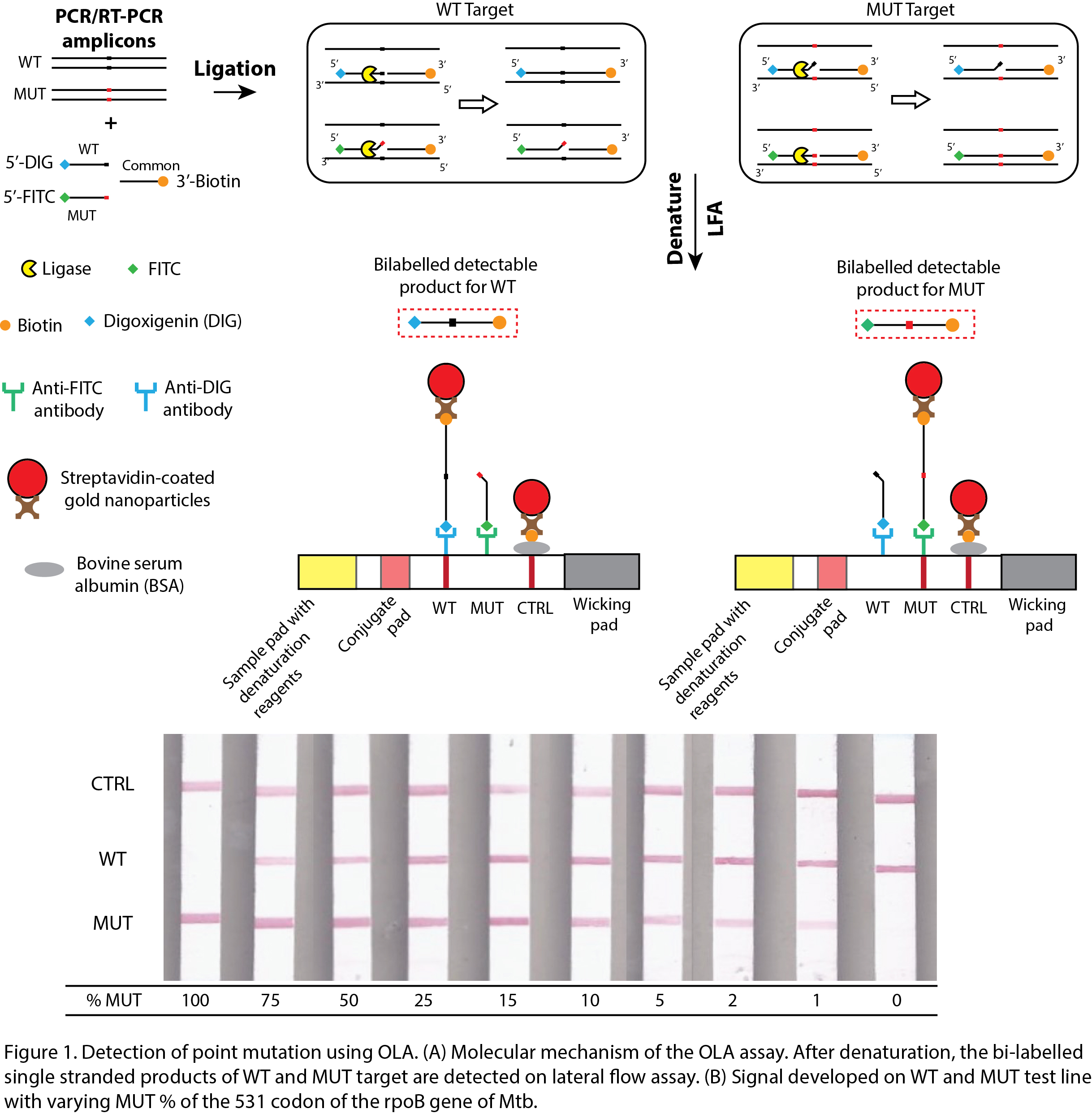
In this work, the point mutations are detected using oligonucleotide assay (OLA). OLA uses three differently labeled oligonucleotides or probes, two of which are allele-specific (Fig. 1A). When the probes hybridize to the amplified DNA, a ligase enzyme stitches the two ends of the probes and establishes a covalent bond (Fig. 1A). The formation of this bond is contingent on the presence or absence of mutation in the target DNA. OLA is a well-established method for SNP-typing but is currently restricted to certified laboratories. It has been used to detect point mutations in HIV using a well-plate-based immunochromatographic assay (Beck et al., 2008, 2014; Panpradist et al., 2016), in the rpoB gene of MTB using “DNA chips†(Jiao-Yu Deng et al., 2004; J.-Y. Deng et al., 2004) and in different genes of human and salmon sperm DNA using fluorescence resonance energy transfer (FRET) (Chen, Livak and Kwok, 1998). However, all these methods require expensive equipment and dedicated infrastructure. We have obviated these requirements by using a low-cost paper-based readout. As the ligated products of OLA are bi-labeled, it can be read on a lateral flow assay (LFA) akin to a home pregnancy strip. LFA consists of two test lines to detect the wild and mutant targets. For homozygous, a single test line would be seen either at WT or MUT, depending on the sample genotype. For heterozygous, relative intensities of the two test lines would indicate the ratio of wild and mutant strain in the sample. Prior to LFA, the ligation mixture is denatured to separate the unligated probes to prevent the generation of false-positive signals on the test lines. Further, dried premixes for PCR and OLA would be developed to reduce pipetting steps and user interventions. The method of deglycerolisation of enzymes radically improves the shelf life of dry-stored enzymes.
A potential application of the assay is in the rapid detection of drug resistance in the tuberculosis-causing bacteria, Mycobacterium tuberculosis (Mtb). Failure to identify specific mutations is the major cause of the spread of resistant strain and the high mortality rate of drug-resistant Tuberculosis (DR-TB) patients. WHO estimated that in 2018 there were half a million new DR-TB cases, majority of which were multi-drug resistant, i.e., resistant to at least rifampicin and isoniazid (first-line drugs for TB diagnosis). Out of these cases, only one-third were reported and diagnosed, highlighting the lack of an appropriate tool for the detection and surveillance of DR-TB. Current genotypic methods to detect DR-TB are NAATs followed by surface hybridization assays (line probe assays (LPAs) and GeneXpert) and real-time PCR assays (using molecular beacon probes). While GeneXpert and LPAs are rapid, their high operational cost restricts their use in certified laboratories. Similarly, real-time PCR requires sophisticated fluorescence imaging instrumentation. OLA, on the contrary, can be performed using low-cost portable PCR machines (e.g., Palm PCRTM and FranklinTM) by minimally trained users, potentially allowing same day selection of appropriate antimicrobial therapy.
Around 96% of mutations causing rifampicin resistance are conferred in the 81 bp rifampicin resistance detection region (RRDR) of the rpoB gene of Mtb. Out of which, the most prevalent mutation is at codon 531: TCG531TTG (S531L). The wild and mutant strains of Mtb having the 531 mutations were differentiated using OLA and LFA in 3 hours. The assay could detect 1 % mutation in a pool of wild and mutant synthetic DNA (Fig. 1B).
While the assay is being developed for DR-TB detection, the methodological and technological reformations witnessed during this project can be adapted to determine drug resistance in any pathogen. The assay thus will be a critical tool for managing infectious diseases in the future.