(49b) Metabolomics Approach to Improving CHO Cell Productivity
AIChE Annual Meeting
2020
2020 Virtual AIChE Annual Meeting
Food, Pharmaceutical & Bioengineering Division
Biomanufacturing: Cell Line Engineering and Omics Studies
Monday, November 16, 2020 - 8:15am to 8:30am
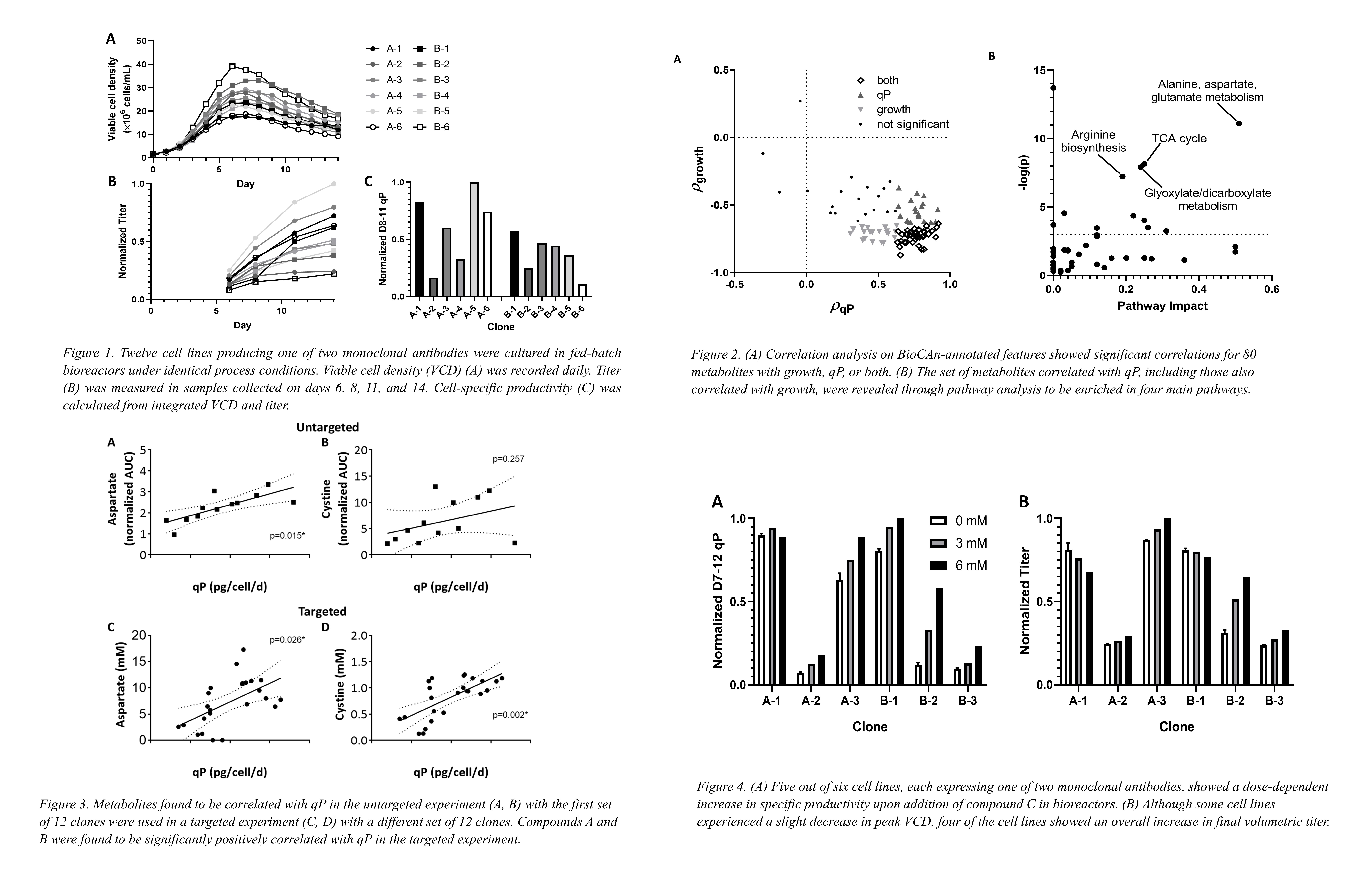
A library of 12 clones from the same host cell line, six producing mAb A and six producing mAb B, was selected based on preliminary data to represent a wide variety of growth and qP profiles. Fed-batch experiments were performed in 5-liter bioreactors using the same production process for all 12 clones, resulting in a range of peak viable cell densities (VCDs), titers and qPs as expected (Fig. 1).
Supernatant samples collected on day 7 (early stationary phase) were analyzed using four different combinations of liquid chromatography (LC) and untargeted mass spectrometry (MS) methods. The MS experiments were performed on a time-of-flight (TOF) LC-MS system (TripleTOF 5600+, AB SCIEX, Framingham, MA). Over 4,000 LC-MS features were detected, including duplicates between methods (i.e., same metabolite detected two or more times by different LC and MS combinations) and adducts. Using a previously developed automated annotation tool (Biologically Consistent Annotation, BioCAn) tool, 99 unique metabolites were putatively identified. Pearson and Spearman correlation coefficients were calculated between each of these putatively identified metabolites and peak VCDs (representing growth) and qPs. Early stationary phase measurements for 80 of these metabolites were significantly correlated (p<0.05) with qP, growth, or both. In general, metabolites significantly correlating with qP had positive associations, while those significantly correlating with growth had negative associations (Fig. 2A). Only 3 metabolites had negative correlations with qP and 1 metabolite had a positive correlation with growth, and none of these were significant. Pathway enrichment analysis using Metaboanalyst (Fig. 2B) showed that metabolites having significant correlations with qP were enriched in alanine, aspartate, and glutamate metabolism. Other enriched pathways included arginine biosynthesis, glyoxylate and dicarboxylate metabolism, and the TCA cycle.
To test whether any of these significantly correlated metabolites could be used as biomarkers of productivity, another fed-batch study was performed with a new library of 12 clones from the same host cell line as above, all producing a third antibody (mAb C). Again, a wide range of growth and productivity profiles was observed. Samples from day 7 of bioreactor culture were collected for a targeted LC-MS experiment (multiple reaction monitoring, MRM). The target analytes comprised a subset of the significantly correlated metabolites identified from the untargeted experiment. In this analysis, compound A was found again to have a significant, positive correlation with qP, confirming an association from the untargeted experiment (Fig. 3A, Fig. 3C). Additionally, compound B, which was positively correlated with qP in the untargeted experiment but did not reach the p<0.05 level of significance, was found in this second set of clones to have a significant, positive correlation (Fig. 3B, Fig. 3D).
Next, a study was performed in fed-batch 250-mL bioreactor cultures to see whether metabolites with significant correlations with qP were directly responsible for improved qP or whether they were byproducts of pathways that are active in a high productivity metabolic state. Select compounds from the enriched pathways were added back as boluses in varying concentrations to cultures of multiple cell lines during mid-exponential phase (day 3). One of these compounds, compound C, showed dose-dependent improvements to qP in five out of the six cell lines tested (Fig. 4A). While the highest dose (6 mM) slightly reduced VCD, overall there was an increase in volumetric titer of 15-107% for four of the cell lines (Fig. 4B).
These studies demonstrate value in using untargeted metabolomics to better understand qP. In this work, we were able to identify potential biomarkers that can be used to shorten cell line development timelines and a metabolite that increased qP and titer when added to bioreactor cultures. Because these studies were conducted across multiple clones, these metabolites and the pathways they belong to may be indicative of a metabolic phenotype that supports a high level of protein production.