(225h) Harnessing Orthogonal tRNA for De Novo Generation of Genetic Codes
AIChE Annual Meeting
2020
2020 Virtual AIChE Annual Meeting
Materials Engineering and Sciences Division
Biomaterials and Life Sciences Engineering: Faculty Candidates III
Wednesday, November 18, 2020 - 9:45am to 10:00am
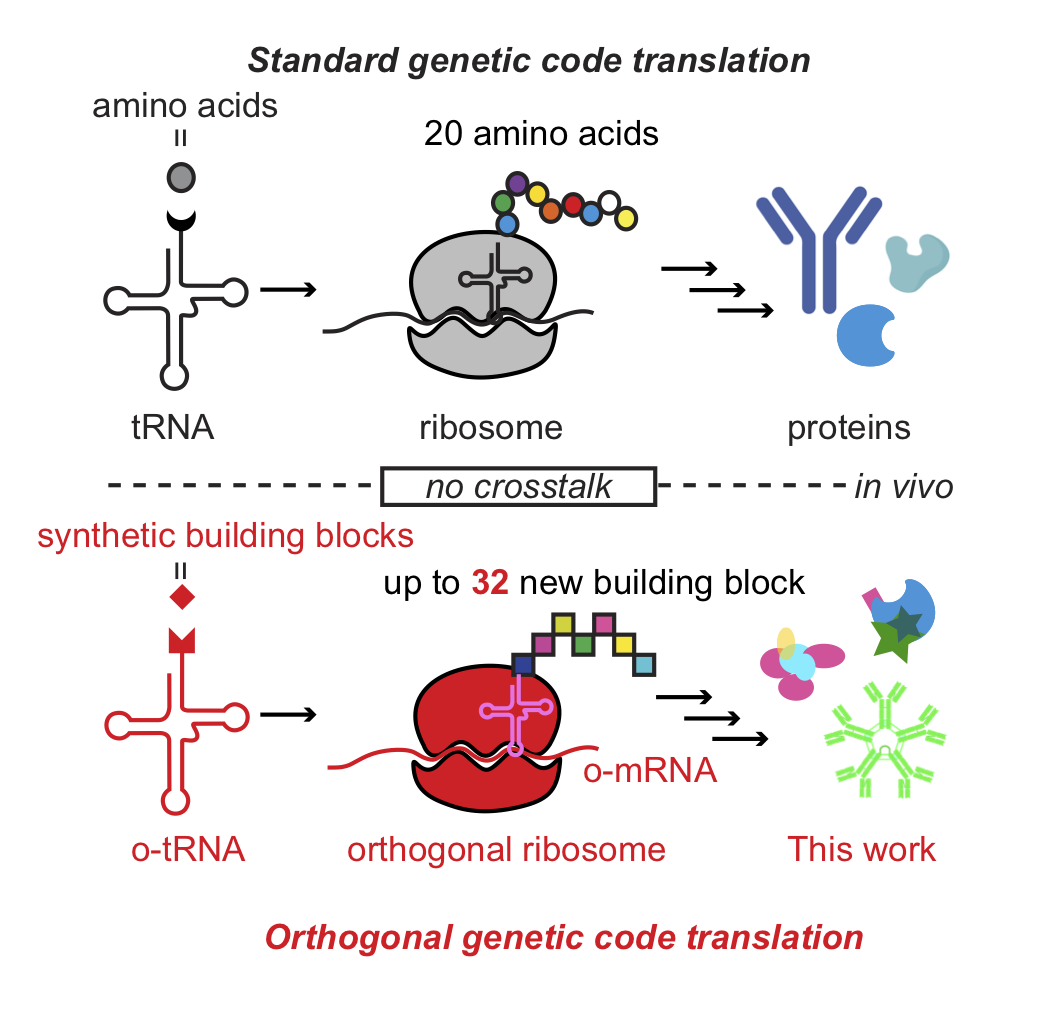
Humanity has managed to leverage the extant ‘standard genetic code’, containing only 20 amino acids, into multi-billion dollar industries. Biomanufactured proteins are used in medicine as therapeutics (e.g. antibodies, insulin), in industrial processes (e.g. production of high fructose corn syrup), and even in the household (e.g. additives to laundry detergents). These examples highlight the extent of functional utility generated from just 20 amino acid building blocks. If we wanted to start mass producing proteins composed of an alternative set of building blocks, we would be faced by the obstacle posed previously: life has evolved and been optimized around using a very specific set of biomolecules and building blocks.
In this talk, I will be discussing strategies, methods, and results for building a parallel genetic code in living organisms. This talk will focus on the development of a generalizable strategy capable of producing >20 mutually orthogonal transfer RNA (o-tRNA). I will be covering challenges and solutions for producing stable o-tRNAs in cells, specifically in E. coli. While o-tRNAs can form the basis for a parallel genetic code, we are faced with the challenge of building a new genetic code from the ground-up. I will then shift focus cover strategies for populating the new codon table in the absence of a functional orthogonal translation system. If successful, orthogonal genetic codes (as described) have the capacity to translate up to 32 additional amino acids or non-standard building blocks. ( Fields area: chemical biology and synthetic biology)