Engineered CRISPR-Cas systems allow scientists to make programmable, precise edits to DNA. This technology has the potential to cure human diseases and transform agriculture.
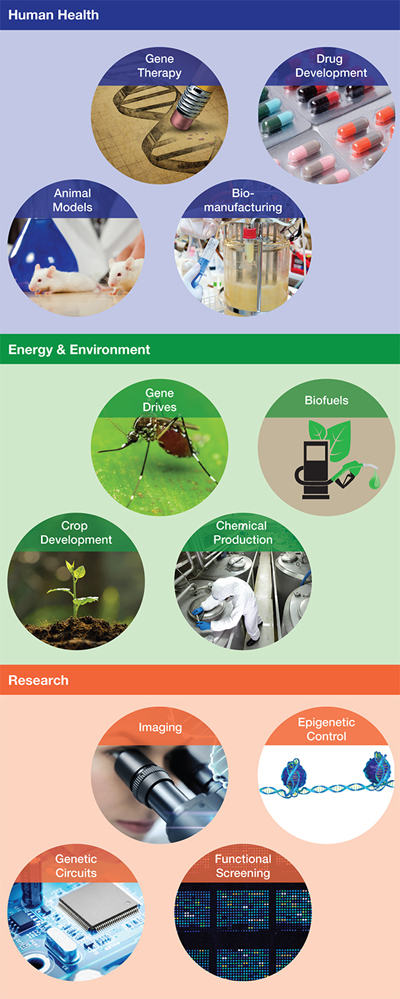
▲Figure 1. CRISPR-Cas technologies have many uses and applications across multiple industries.
CRISPR-Cas9 has emerged as a powerful new gene-editing tool, giving researchers the power to quickly and easily alter DNA. With applications from gene therapy to crop development, genome engineering has the potential to cure disease and solve world hunger (Figure 1). These capabilities have also raised major ethical questions that are being actively debated worldwide.
Here, we explain what CRISPR is, how it works, why it’s revolutionizing biotechnology, and the multiple debates throughout society that have been catalyzed by the technology. This article is meant to serve as a primer; many excellent reviews offer greater depth on the natural role of CRISPR (1–3) and its many technological uses (4–7).
CRISPR-Cas systems are bacterial adaptive immune systems
CRISPR (clustered regularly interspaced short palindromic repeats) technologies have their origins in the prokaryotic world of bacteria and archaea. Like humans, bacteria have their own type of viruses. When these viruses infect a cell, they inject their DNA in an attempt to co-opt the cellular host machinery to produce more copies of the virus. Phage proliferation often leaves behind a damaged or dead bacterial host cell. As a result, bacteria have evolved multilayered antiviral defense systems. CRISPR-Cas systems are highly adaptive and inheritable systems that represent one mechanism of prokaryotic defense. CRISPR-Cas systems are unique in their ability to create memory in reaction to an initial infection, which enhances immune response to subsequent reinfections.
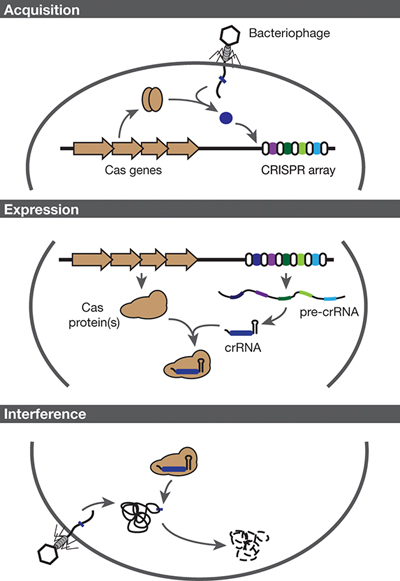
▲Figure 2. CRISPR-Cas systems provide bacteria with adaptive immunity. Immunity is conferred through three steps: acquisition, expression, and interference. Acquisition: Cas proteins incorporate a small piece of foreign DNA as a new spacer within the CRISPR array. Expression: The CRISPR array is transcribed and undergoes processing by the Cas protein(s) and accessory factors to form the CRISPR RNA (crRNA). Interference: The effector complex, comprised of the crRNA and Cas protein(s), targets and degrades invading DNA.
CRISPR-Cas systems provide immunity through a general three-step pathway: acquisition, expression, and interference (Figure 2, pg. 38). During the first stage, foreign genetic material infects the cell. Cas proteins recognize this viral DNA (or RNA) and generate fragments of that viral DNA to be copied and pasted into the bacterial genome at the CRISPR locus. This sampling of viral DNA is what drives the adaptive element of the immune response. In this manner, the CRISPR-Cas system allows the bacteria to retain memory of past infections.
During the second stage of immunity, the cell expresses the CRISPR locus and produces CRISPR RNA (crRNA) molecules, which contain an exact replica of the previously seen viral DNA. The crRNAs work together with Cas proteins and accessory factors to protect the cells. Because RNA is a chemical cousin of DNA, it can interact with DNA molecules that have a complementary sequence.
In the final stage, a phage reinfects the bacterium. This time, however, the protein-crRNA effector complex recognizes the viral DNA that matches the crRNA sequence. This recognition triggers the complex to degrade and eliminate the viral DNA, thereby protecting the cell.
While immunity is the natural function of CRISPR-Cas systems, researchers discovered that these effector complexes are programmable, so scientists can engineer these systems to recognize not only viral DNA, but also almost any DNA sequence of our choosing. It is this programmable recognition of nucleic acids that makes CRISPR so appealing as a basis for genetic tools.
Exploiting the power of CRISPR-Cas systems
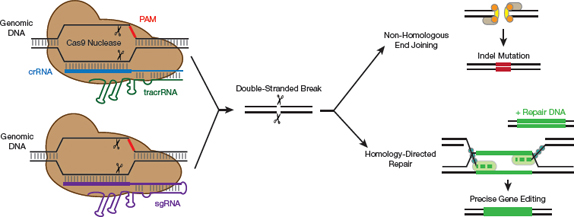
▲Figure 3. The CRISPR-Cas9 mechanism of action. Cas9 naturally relies on two pieces of short RNA: crRNA and tracrRNA. The engineered sgRNA combines these two elements into a single RNA molecule. In order to find the target, Cas9 scans the DNA for a sequence termed the protospacer adjacent motif (PAM). The Cas9 then unzips the DNA and determines whether the sgRNA matches the target DNA. If they do match, the Cas9 generates a double-stranded DNA break. When this occurs, the cell tries to repair the cut.
While there are many different types of CRISPR-Cas systems, the CRISPR craze largely centers around one specific protein, Cas9. To function, Cas9 must be complexed with a crRNA and a trans-activating crRNA (tracrRNA). This has been simplified by combining the crRNA and the tracrRNA into an engineered single-guide RNA (sgRNA) (8). As a result, only two components — the Cas9 protein and the sgRNA molecule — are required for this engineered system to function (Figure 3, pg. 39).
To recognize a target sequence, Cas9 first searches for a short conserved sequence called the protospacer adjacent motif (PAM) (9). Once Cas9 identifies a PAM, it then checks whether the target and the sgRNA are complementary. If the sequence is confirmed as a match, Cas9 cuts the target DNA, generating a blunt-ended double-stranded break.
The double-stranded break alone does not result in editing. Editing events occur when the cell tries to repair the damage via either non-homologous end-joining or homology-directed repair (10).
The non-homologous end-joining repair process is often error-prone. In an attempt to mend the gap in the DNA, the repair machinery often generates small insertions or deletions in the affected area. These random mutations can inactivate genes, which can allow researchers to understand gene function.
Although knocking out genes in this way enables researchers to identify gene function, sometimes changes need to be better defined. Homology-directed repair can be used to generate more systematic changes. In this technique, a piece of...
Would you like to access the complete CEP Article?
No problem. You just have to complete the following steps.
You have completed 0 of 2 steps.
-
Log in
You must be logged in to view this content. Log in now.
-
AIChE Membership
You must be an AIChE member to view this article. Join now.
Copyright Permissions
Would you like to reuse content from CEP Magazine? It’s easy to request permission to reuse content. Simply click here to connect instantly to licensing services, where you can choose from a list of options regarding how you would like to reuse the desired content and complete the transaction.
